Journal:
Molecular Cell. 2018 December; Vol 72, p.1–13
Authors:
Manpreet Kalkat, Diana Resetca, Corey Lourenco, Pak-Kei Chan, Yong Wei, Yu-Jia Shiah, Natasha Vitkin, Yufeng Tong, Maria Sunnerhagen, Susan J. Done, Paul C. Boutros, Brian Raught and Linda Z. Penn .
Abstract:
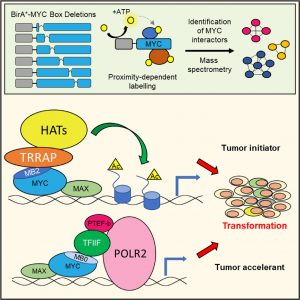
Transforming members of the MYC family (MYC, MYCL1, and MYCN) encode transcription factors containing six highly conserved regions, termed MYC homology boxes (MBs). By conducting proteomic profiling of the MB interactomes, we demonstrate that half of the MYC interactors require one or more MBs for binding. Comprehensive phenotypic analyses reveal that two MBs, MB0 and MBII, are universally required for transformation. MBII mediates interactions with acetyltransferase-containing complexes, enabling histone acetylation, and is essential for MYC-dependent tumor initiation. By contrast, MB0 mediates interactions with transcription elongation factors via direct binding to the general transcription factor TFIIF. MB0 is dispensable for tumor initiation but is a major accelerator of tumor growth. Notably, the full transforming activity of MYC can be restored by co-expression of the non-transforming MB0 and MBII deletion proteins, indicating that these two regions confer separate molecular functions, both of which are required for oncogenic MYC activity.
Link: https://www.cell.com/molecular-cell/fulltext/S1097-2765(18)30802-5#%20
DOI: 10.1016/j.molcel.2018.09.031


